import matplotlib.pyplot as plt
from matplotlib import font_manager as fm
import numpy as np
font_size = 22.5
colwidth = 9.12103352 # for figsize width
mm_to_pt = 1/25.4*72
linewidth = 1.6141975081451896 * mm_to_pt # mm
col_red = '#C00A35'
col_gold = '#FFCD00'
col_orange = '#F3965E'
col_plum = '#AA1555'
col_green = '#24A793'
col_title = '#000000'
plt.rcParams['font.family'] = 'Open Sans'
plt.rcParams['font.style'] = 'normal'
plt.rcParams['font.weight'] = 'normal'
plt.rcParams.update({'figure.autolayout': True})
np.random.seed(42)
golden = (1 + 5 ** 0.5) / 2
# Plot heatmaps for each channel
sampling_rate = 2048In [1]:
In [19]:
import matplotlib.pyplot as plt
import numpy as np
from matplotlib import font_manager as fm
font_size = 22.5
colwidth = 9.12103352
mm_to_pt = 1/25.4*72
linewidth = 1.6141975081451896 * mm_to_pt # mm
col_gold = '#ffcd00'
# Ensure Open Sans font is available
# font_path = fm.findSystemFonts(fontpaths=None, fontext='ttf')
# open_sans = [path for path in font_path if 'OpenSans-Regular.ttf' in path]
# prop = fm.FontProperties(fname=open_sans[0])
# if open_sans:
plt.rcParams['font.family'] = 'Open Sans'
plt.rcParams['font.style'] = 'normal'
plt.rcParams['font.weight'] = 'normal'
plt.rcParams.update({'figure.autolayout': True})
# plt.rcParams['font.family'] = prop.get_family()
# else:
# print("Open Sans font not found. Using default font.")
# plt.figure(figsize=(9.12103352, 8))
# Create figure and axes
fig, ax = plt.subplots(2, 1, figsize=(colwidth, 3), gridspec_kw={'height_ratios': [1, 1]})
# Plot EEG response for traditional flickering stimuli
t = np.arange(0, 100, 1)
flicker_response = 0.5 * (1 + 0.5 * np.sin(2 * np.pi * 5 * t / 100))
ax[0].plot(t, flicker_response, label='Visible Flicker', color=col_gold, linewidth=linewidth, clip_on=False)
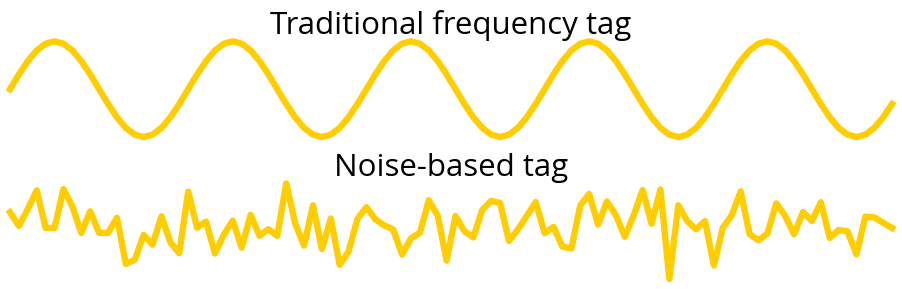
ax[0].set_title('Traditional frequency tag', fontsize=font_size)
ax[0].axis('off')
ax[0].margins(0)
# Plot EEG response for noise-based tagging
np.random.seed(42)
noise_response = np.random.normal(0, 1, 100)
ax[1].plot(t, noise_response, label='High-Frequency Noise', color=col_gold, linewidth=linewidth, clip_on=False)
ax[1].set_title('Noise-based tag', fontsize=font_size)
ax[1].axis('off')
ax[1].margins(0)
plt.subplots_adjust(left=0, right=1,
# top=1, bottom=0
# hspace=.3
)
plt.tight_layout()
# Title and layout adjustments
# plt.suptitle('Comparison of Traditional and Noise-Based Tagging in BCIs', fontsize=14)
plt.savefig('../presentations/figures/tagging_comparison.svg', bbox_inches=None, pad_inches=None, transparent=True)
plt.show()In [20]:
import matplotlib.pyplot as plt
import matplotlib.patches as patches
import numpy as np
col_orange = '#F3965E'
col_plum = '#AA1555'
col_green = '#24A793'
col_red = '#C00A35'
# col_title = '#3b3b3b'
col_title = '#000000'
def create_trial_timeline():
# Constants based on the experiment code
cue_duration = 0.15
stim_delay = 1.0
stim_duration = 1.0
target_duration = 0.1
response_duration = 3.0
# Time calculations
pre_trial_start = -1.0
cue_onset = 0.0
cue_offset = cue_onset + cue_duration
stim_onset = cue_offset + stim_delay
stim_offset = stim_onset + stim_duration
target_onset_min = stim_onset + 0.2
target_onset_max = stim_onset + 0.8
np.random.seed(42)
target_onset = stim_onset + np.random.uniform(0.2, 0.8)
target_offset = target_onset + target_duration
response_allowance = stim_offset + response_duration
end_time = response_allowance + 0
# Define phases
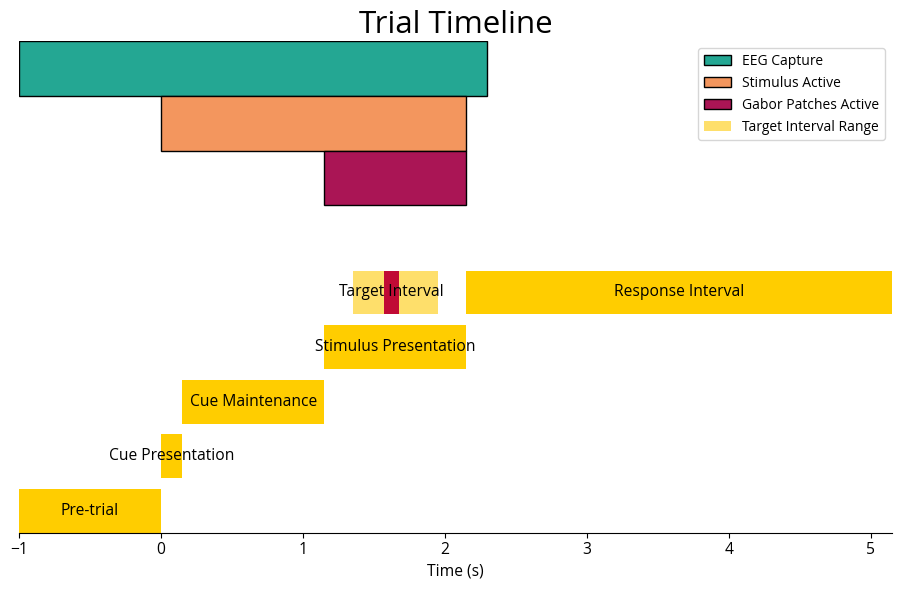
phases = [
('Pre-trial', pre_trial_start, cue_onset),
('Cue Presentation', cue_onset, cue_offset),
('Cue Maintenance', cue_offset, stim_onset),
('Stimulus Presentation', stim_onset, stim_offset),
# ('Target Interval', target_onset, target_offset),
('Response Interval', stim_offset, response_allowance)
]
fig, ax = plt.subplots(figsize=(colwidth, 6), clip_on=False)
# Add overarching bars
ax.add_patch(patches.Rectangle((pre_trial_start, 4.0), 2.3 - pre_trial_start, 0.5, facecolor=col_green, edgecolor=col_title, label='EEG Capture'))
ax.add_patch(patches.Rectangle((cue_onset, 3.5), stim_offset - cue_onset, 0.5, facecolor=col_orange, edgecolor=col_title, label='Stimulus Active'))
ax.add_patch(patches.Rectangle((stim_onset, 3.0), stim_offset - stim_onset, 0.5, facecolor=col_plum, edgecolor=col_title, label='Gabor Patches Active'))
# Plot phases
for idx, (label, start, end) in enumerate(phases):
ax.broken_barh([(start, end - start)], (idx * 0.5, 0.4), facecolors=(col_gold))
ax.text((start + end) / 2, idx * 0.5 + 0.2, label, ha='center', va='center', color='black', fontsize=font_size/2)
# Add the target interval range as a shaded area
ax.add_patch(patches.Rectangle((target_onset_min, 2.0), target_onset_max - target_onset_min, 0.4, facecolor='#fedf6b', alpha=1, label='Target Interval Range'))
# Add the actual target interval
ax.broken_barh([(target_onset, target_duration)], (2.0, 0.4), facecolors=(col_red))
ax.text(target_onset + target_duration / 2, 2.0 + 0.2, 'Target Interval', ha='center', va='center', color='black', fontsize=font_size*.5)
# Add legend
ax.legend(loc='upper right')
# Set labels and title
ax.set_yticks([])
ax.set_xlabel('Time (s)', fontsize=font_size/2, color=col_title)
ax.set_xlim(pre_trial_start, end_time)
ax.tick_params(axis='x', labelsize=font_size/2)
ax.set_title('Trial Timeline', fontsize=font_size, color=col_title)
ax.spines[['right', 'left', 'top']].set_visible(False)
ax.margins(0)
plt.subplots_adjust(left=0, right=1,
#top=1, bottom=0
)
plt.savefig('../presentations/figures/trial_timeline.svg', bbox_inches=None, pad_inches=None, transparent=True)
plt.show()
# Call the function to create the plot
create_trial_timeline()In [21]:
from concurrent.futures import ProcessPoolExecutor, as_completed
import h5py
import matplotlib.cm as cm
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import scipy.io as sio
import seaborn as sns
from scipy.interpolate import interp1d
from scipy.signal import (butter, correlate, filtfilt, find_peaks, resample,
sosfiltfilt, welch)
trial = 'A2'
eeg_filepath = f'../data/{trial}/converted/eeg_matrix.mat'
eeg_trialinfo = f'../data/{trial}/converted/trialinfo_matrix_{trial}_cleanedtrials.mat'
eeg_raw_data = f'../data/{trial}/experimental/data.mat'
eeg_labels_path = f'../data/A0/preprocessing/channel_labels.mat' # always get the labels from A0
f = sio.loadmat(eeg_labels_path)
labels = f['channellabels_ADSselection']
def load_mat_file(filepath):
""" Load a MATLAB .mat file and return its content. """
try:
mat_contents = sio.loadmat(filepath)
return mat_contents
except NotImplementedError:
# If the file is v7.3, it will need h5py to handle it
with h5py.File(filepath, 'r') as file:
return {key: np.array(value) for key, value in file.items()}
def get_eeg_data(eeg_data_path):
""" Extract EEG data from the .mat file. """
data = load_mat_file(eeg_data_path)
eeg_data = data['data_eeg']
return eeg_data
def get_trial_info(trial_info_path):
""" Extract trial information from the .mat file. """
data = load_mat_file(trial_info_path)
trial_info = data['all_info']
return trial_info
def load_raw_data(noise_data_path):
""" Load and return the entire noise stimuli structure from the .mat file. """
mat_contents = load_mat_file(noise_data_path)
# Extract the 'noise_stims' from the nested structure
noise_stims = mat_contents['data'][0, 0]
return noise_stims
def get_specific_raw_data(idx, eeg_raw_data, block_index, trial_index):
""" Fetch the specific noise stimulus for the given block, trial, and channel (0 for left, 1 for right). """
idxs = ['reaction_times', 'reaction_err', 'answers', 'base_delay', 'target_timings', 'flicker_sides', 'attend_sides', 'orients_L', 'orients_R', 'angle_magnitude', 'probe_sides', 'missed', 'targets_binary', 'tagging_types', 'noise_stims', 'p_num', 'date']
return eeg_raw_data[idxs.index(idx)][block_index, trial_index]
def trial_number_to_indices(trial_number):
""" Convert trial number to block and trial indices, zero-indexed. """
trial_number = int(trial_number) - 1
block_index = trial_number // 32 + 1 # +1 block because we've discard the first block
trial_index = trial_number % 32
return block_index, trial_index
def load_and_process_data(eeg_filepath, trialinfo_filepath, raw_data_filepath):
eeg_data = get_eeg_data(eeg_filepath)
trial_info = get_trial_info(trialinfo_filepath)
eeg_raw_data = load_raw_data(raw_data_filepath)
processed_trials = []
# Assuming we loop over actual trial data, adjust as necessary
for trial, eeg_data in zip(trial_info, eeg_data):
block_index, trial_index = trial_number_to_indices(trial[14])
# Access the corresponding noise stimulus for left and right channels
noise_stim_left, noise_stim_right = get_specific_raw_data('noise_stims', eeg_raw_data, block_index, trial_index)
trial_data = {
'eeg': eeg_data, # Adjust for zero-index and skip block 1
'trial_info': trial,
'reaction_time_1': trial[2],
'reaction_time_2': get_specific_raw_data('reaction_times', eeg_raw_data, block_index, trial_index),
'has_noise': bool(int(trial[13])),
'has_56|64': int(trial[7]) == 0,
'has_56|60': int(trial[7]) == 1,
'attended_side': trial[8],
'noise_stim_left': noise_stim_left,
'noise_stim_right': noise_stim_right,
'base_delay': trial[6],
'target_timings': trial[5],
}
processed_trials.append(trial_data)
return processed_trials
experiment_data = load_and_process_data(eeg_filepath, eeg_trialinfo, eeg_raw_data)
experiment_data = pd.DataFrame(experiment_data)
# These are in the form trials (440), samples (6758), channels (37)
eeg_data = np.stack(experiment_data['eeg'])
channel_labels = load_mat_file(eeg_labels_path)['channellabels_ADSselection'].flatten()
channel_labels = [x[0] for x in channel_labels][:-4] # remove the last 4 channels, they're not used (EXG1 EXG2 EXG3 EXG4)
print(' '.join(channel_labels))Fp1 AF7 AF3 F1 F3 F5 F7 FT7 FC5 FC3 FC1 C1 C3 C5 T7 TP7 CP5 CP3 CP1 P1 P3 P5 P7 PO7 PO3 O1 Iz Oz POz Pz P2 P4 P6 P8 PO8 PO4 O2In [22]:
def butter_bandpass(lowcut, highcut, fs, order=5):
nyquist = 0.5 * fs
low = lowcut / nyquist
high = highcut / nyquist
b, a = butter(order, [low, high], btype='band')
return b, a
def bandpass_filter(data, lowcut, highcut, fs, order=5):
b, a = butter_bandpass(lowcut, highcut, fs, order=order)
y = filtfilt(b, a, data)
return y
def butter_bandpass_sos(lowcut, highcut, fs, order=5):
nyquist = 0.5 * fs
low = lowcut / nyquist
high = highcut / nyquist
sos = butter(order, [low, high], btype='band', output='sos')
return sos
def bandpass_filter_sos(data, lowcut, highcut, fs, order=5):
sos = butter_bandpass_sos(lowcut, highcut, fs, order=order)
y = sosfiltfilt(sos, data)
return y
def mix_signals(eeg_data, noise_signal, strength):
mixed_data = (1-strength) * eeg_data + strength * noise_signal
return mixed_data
def upsample_or_downsample(data, original_freq, new_freq):
""" Upsample or downsample data to the new frequency. """
num_samples = int(len(data) * new_freq / original_freq)
return resample(data, num_samples)
def normalise_signal(signal):
return (signal - np.mean(signal)) / np.std(signal)
def compute_best_combined_offset(eeg_data, noise_stim_left, noise_stim_right, channel_labels,
sampling_rate=2048, noise_freq=480,
upsample_noise=True, eeg_start_time=0.0, eeg_end_time=3.2,
noise_start_time=0.0, noise_end_time=2.0,
bandpass_eeg=None, bandpass_noise=None, mix_noise_strength=None, plot_results=False):
num_samples, num_channels = eeg_data.shape
# num_channels = len(channel_labels)
eeg_segment_start = int(eeg_start_time * sampling_rate)
eeg_segment_end = int(eeg_end_time * sampling_rate)
# Select EEG segment
eeg_data_segment = eeg_data[eeg_segment_start:eeg_segment_end, :].copy()
# Select noise segment
noise_segment_start = int(noise_start_time * noise_freq)
noise_segment_end = int(noise_end_time * noise_freq)
noise_stim_left_segment = noise_stim_left[noise_segment_start:noise_segment_end].copy()
noise_stim_right_segment = noise_stim_right[noise_segment_start:noise_segment_end].copy()
# Optionally bandpass filter the signals
if bandpass_eeg:
lowcut, highcut = bandpass_eeg
for channel in range(num_channels):
eeg_data_segment[:, channel] = bandpass_filter_sos(eeg_data_segment[:, channel], lowcut, highcut, sampling_rate)
if bandpass_noise:
lowcut, highcut = bandpass_noise
noise_stim_left_segment = bandpass_filter_sos(noise_stim_left_segment, lowcut, highcut, noise_freq)
noise_stim_right_segment = bandpass_filter_sos(noise_stim_right_segment, lowcut, highcut, noise_freq)
# Upsample or downsample the noise signals
if upsample_noise:
noise_stim_left_segment = upsample_or_downsample(noise_stim_left_segment, noise_freq, sampling_rate)
noise_stim_right_segment = upsample_or_downsample(noise_stim_right_segment, noise_freq, sampling_rate)
else:
eeg_data_segment = upsample_or_downsample(eeg_data_segment, sampling_rate, noise_freq)
sampling_rate = noise_freq
# Normalise the signals before mixing and cross-correlation
noise_stim_left_segment = normalise_signal(noise_stim_left_segment)
noise_stim_right_segment = normalise_signal(noise_stim_right_segment)
eeg_data_segment = normalise_signal(eeg_data_segment)
# Optionally merge the noise signals
mixed_noise_signal = (noise_stim_left_segment + noise_stim_right_segment) / 2
# Optionally mix the noise signal into the EEG data
if mix_noise_strength is not None:
offset = 1 * sampling_rate
for channel in range(num_channels):
eeg_data_segment[offset:offset+len(mixed_noise_signal), channel] = mix_signals(eeg_data_segment[offset:offset+len(mixed_noise_signal), channel], mixed_noise_signal, mix_noise_strength)
# Store the cross-correlation results and lags
cross_corr_results = []
lags = []
max_cors = []
for channel, channel_label in zip(range(num_channels), channel_labels):
# if channel_label not in ['Pz', 'POz', 'Iz', 'O1', 'PO3']: continue # Only plot the interesting occipital channels
eeg_channel_data = eeg_data_segment[:, channel]
# Compute cross-correlation with the mixed noise signal
corr = correlate(eeg_channel_data, mixed_noise_signal, mode='full')[len(mixed_noise_signal) - 1:
len(mixed_noise_signal) - 1 + len(eeg_channel_data)]
# Above aligns corr, so that the measured lag corresponds directly to where the signal starts in the EEG data
# Find the peak in the cross-correlation
peak_lag = np.argmax(corr)
# Store results
cross_corr_results.append(corr)
lags.append(peak_lag)
max_cors.append(np.max(corr))
# lags = [2048]
# print('Max Cross-Correlation:', max_cors)
# print('Lags:', sorted(lags))
# print('Sorted:')
# for tup in sorted(zip(channel_labels, lags, max_cors), key=lambda x: x[1]):
# print(tup)
if not plot_results:
return lags, cross_corr_results
# Plotting the results
time_vector = np.linspace(eeg_start_time, eeg_end_time, eeg_data_segment.shape[0])
for channel in range(num_channels):
plt.figure(figsize=(40, 10)) # Adjusting the figure size for better visualization
normalized_eeg = normalise_signal(eeg_data_segment[:, channel])
normalized_noise = normalise_signal(mixed_noise_signal)
normalized_corr = normalise_signal(cross_corr_results[channel])
print(len(normalized_eeg), len(normalized_noise), len(normalized_corr))
peak_time = lags[channel] / sampling_rate
offset_time_vector = np.linspace(0, len(normalized_noise) / sampling_rate, len(normalized_noise)) + peak_time
# Plot cross-correlation in the upper subplot
ax1 = plt.subplot(2, 1, 1)
plt.plot(time_vector, normalized_corr, color='g', label='Cross-Correlation', alpha=0.7, linestyle='-')
plt.axvline(x=peak_time, color='r', linestyle='--', label='Detected Peak')
plt.legend()
plt.title(f'Cross-Correlation for Channel {channel + 1} / {channel_labels[channel]}')
plt.xlabel('Time (seconds)')
plt.ylabel('Normalized Correlation')
plt.xlim(eeg_start_time, eeg_end_time)
# Plot EEG and noise signals in the lower subplot
ax2 = plt.subplot(2, 1, 2)
plt.plot(time_vector, normalized_eeg, label=f'EEG Channel {channel + 1}', alpha=0.7)
plt.plot(offset_time_vector, normalized_noise, label='Offset Noise Signal', alpha=0.7)
plt.axvline(x=peak_time, color='r', linestyle='--', label='Detected Peak')
plt.legend()
plt.title(f'EEG, Offset Noise, and Cross-Correlation for Channel {channel + 1} / {channel_labels[channel]}')
plt.xlabel('Time (seconds)')
plt.ylabel('Normalized Amplitude')
plt.xlim(eeg_start_time, eeg_end_time)
plt.show()
return lags, cross_corr_results
def process_experiment(experiment, channel_labels):
eeg_data = experiment['eeg']
noise_stim_left = experiment['noise_stim_left']
noise_stim_right = experiment['noise_stim_right']
lags, correlations = compute_best_combined_offset(eeg_data, noise_stim_left, noise_stim_right, channel_labels,
upsample_noise=True,
eeg_start_time=0.0, eeg_end_time=3.2,
noise_start_time=0.0, noise_end_time=2.0, # noise times present in the experiment
# noise_start_time=4.0, noise_end_time=6.0, # noise times not present in the experiment
bandpass_eeg=(50, 80), bandpass_noise=(50, 80),
# mix_noise_strength=0.2,
plot_results=False)
# correlations = [interpolate_local_maxima(corr, 2048)[0] for corr in correlations]
return correlations
def parallel_process_experiments(experiments, channel_labels, num_channels):
cross_corrs_all_trials = [[] for _ in range(num_channels)]
with ProcessPoolExecutor() as executor:
futures = {executor.submit(process_experiment, experiment, channel_labels): i for i, experiment in experiments.iterrows()}
for future in as_completed(futures):
correlations = future.result()
for channel in range(num_channels):
cross_corrs_all_trials[channel].append(correlations[channel])
return cross_corrs_all_trials
num_channels = 37
noise_experiments = experiment_data[experiment_data['has_noise']]
cross_corrs_all_trials = parallel_process_experiments(noise_experiments, channel_labels, num_channels)
# Plot heatmaps for each channel
sampling_rate = 2048In [23]:
import mne
def calculate_snr(cross_corrs_all_trials, sampling_rate, expected_peak_time=None):
snrs = []
for channel_corrs in cross_corrs_all_trials:
cumulative_corr = np.sum(channel_corrs, axis=0)
# Time vector for the cross-correlation
# time_vector = np.linspace(0, len(cumulative_corr) / sampling_rate, len(cumulative_corr))
if expected_peak_time is not None:
# Find the peak closest to the expected peak time
peak_signal = np.max(cumulative_corr[int((expected_peak_time - 0.0)*sampling_rate):
int((expected_peak_time + 0.1)*sampling_rate)])
else:
# Find the highest peak in the cross-correlation
print(np.max(cumulative_corr))
peak_signal = np.max(cumulative_corr)
# Calculate the noise as the standard deviation of the cross-correlation
noise = np.std(cumulative_corr) # Assuming this represents the noise
snr = 10 * np.log10(peak_signal**2 / noise**2)
snrs.append(snr)
return snrs
# Plot topography
def plot_topography(snr_values, channel_labels, sampling_rate):
info = mne.create_info(ch_names=channel_labels, sfreq=sampling_rate, ch_types='eeg')
# Use a standard montage for electrode positions
montage = mne.channels.make_standard_montage('biosemi64')
info.set_montage(montage)
# Create Evoked object with SNR values
evoked_data = np.array(snr_values).reshape(-1, 1)
evoked = mne.EvokedArray(evoked_data, info)
# Plot topography
fig, ax = plt.subplots(figsize=(colwidth, 4))
im, cn = mne.viz.plot_topomap(evoked.data[:, 0], evoked.info, axes=ax, show=False)
ax.margins(0)
cbar = fig.colorbar(im, ax=ax, format="%0.1f dB")
cbar.ax.tick_params(labelsize=font_size/2)
plt.subplots_adjust(left=0, right=1,
top=1, bottom=0
)
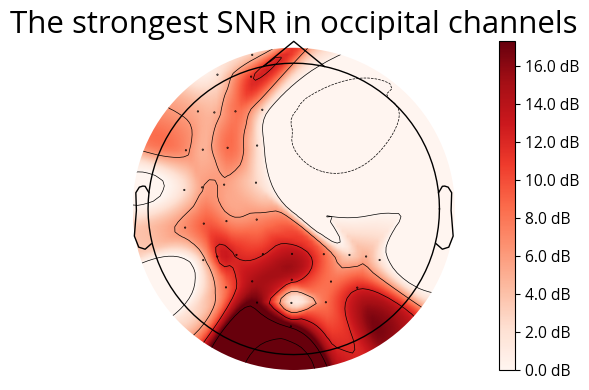
ax.set_title('The strongest SNR in occipital channels', fontsize=font_size)
plt.savefig('../presentations/figures/snr_topography.svg', bbox_inches='tight', pad_inches=None, transparent=True)
plt.show()
# Example usage
expected_peak_time = 1.077 # Adjust based on where you expect the peak
snr_values = calculate_snr(cross_corrs_all_trials, sampling_rate, expected_peak_time)
plot_topography(snr_values, channel_labels, sampling_rate)
print(snr_values, channel_labels, sampling_rate)[np.float64(4.029172557196618), np.float64(2.2400762736479014), np.float64(11.695039968214257), np.float64(2.1354880525172684), np.float64(8.209536805498358), np.float64(4.917421652932336), np.float64(4.882090722896512), np.float64(6.493174921918885), np.float64(4.760766207602362), np.float64(8.469544107179415), np.float64(1.9680384920347498), np.float64(2.5825144827207556), np.float64(5.668645455147909), np.float64(5.915159436186508), np.float64(2.624023920921377), np.float64(7.055892579861621), np.float64(9.30160822622965), np.float64(7.040518549383671), np.float64(6.970904052445302), np.float64(11.814415498470403), np.float64(11.812828959531974), np.float64(12.248904620395486), np.float64(4.939831549275856), np.float64(9.04594248704358), np.float64(14.508447212003306), np.float64(16.78457176599062), np.float64(17.287311672049633), np.float64(1.2760884477553238), np.float64(13.26572313322318), np.float64(13.851556791322249), np.float64(8.758936750362473), np.float64(2.693660896419111), np.float64(4.825748911926802), np.float64(1.9825153904516102), np.float64(11.61165031241092), np.float64(8.159933499133336), np.float64(8.333717016157046)] [np.str_('Fp1'), np.str_('AF7'), np.str_('AF3'), np.str_('F1'), np.str_('F3'), np.str_('F5'), np.str_('F7'), np.str_('FT7'), np.str_('FC5'), np.str_('FC3'), np.str_('FC1'), np.str_('C1'), np.str_('C3'), np.str_('C5'), np.str_('T7'), np.str_('TP7'), np.str_('CP5'), np.str_('CP3'), np.str_('CP1'), np.str_('P1'), np.str_('P3'), np.str_('P5'), np.str_('P7'), np.str_('PO7'), np.str_('PO3'), np.str_('O1'), np.str_('Iz'), np.str_('Oz'), np.str_('POz'), np.str_('Pz'), np.str_('P2'), np.str_('P4'), np.str_('P6'), np.str_('P8'), np.str_('PO8'), np.str_('PO4'), np.str_('O2')] 2048In [24]:
cross_corrs_all_trials = parallel_process_experiments(noise_experiments, channel_labels, num_channels)
golden = (1 + 5 ** 0.5) / 2
# Plot heatmaps for each channel
sampling_rate = 2048
for channel, channel_label in enumerate(channel_labels):
if channel_label not in ['Iz']: continue # Only plot the interesting occipital channels
fig, ax = plt.subplots(figsize=(colwidth, 3), clip_on=False)
ax.spines[['right', 'left', 'top']].set_visible(False)
# Plot heatmap of cross-correlations for all trials
# plt.subplot(2, 1, 1)
# sns.heatmap(np.array(cross_corrs_all_trials[channel]), cmap='viridis', cbar=False)
# plt.title(f'Heatmap of Cross-Correlations for Channel {channel + 1} / {channel_labels[channel]}')
# plt.xlabel('Lag')
# plt.ylabel('Trial')
# plt.subplot(2, 1, 2)
# Calculate and plot cumulative cross-correlation
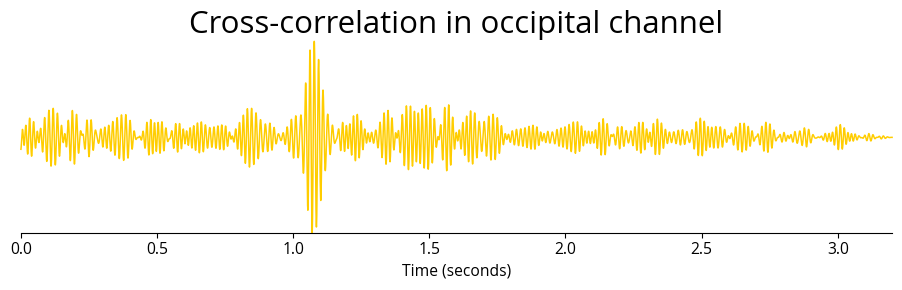
cumulative_corr = np.sum(np.array(cross_corrs_all_trials[channel]), axis=0)
time_vector = np.linspace(0, len(cumulative_corr) / sampling_rate, len(cumulative_corr))
plt.plot(time_vector, cumulative_corr, color=col_gold, linewidth=linewidth/golden**3, clip_on=False)
plt.title(f'Cross-correlation in occipital channel', fontsize=font_size)
plt.xlabel('Time (seconds)', fontsize=font_size/2)
plt.xticks(fontsize=font_size/2)
# plt.ylabel('Cumulative Correlation')
# remove y axis
ax.yaxis.set_visible(False)
ax.margins(0)
plt.xlim(0, time_vector[-1])
# plt.legend()
plt.subplots_adjust(left=0, right=1,
# top=1, bottom=0
)
plt.tight_layout()
plt.savefig(f'../presentations/figures/cross_correlation_{channel_label}.svg', bbox_inches=None, pad_inches=None, transparent=True)
plt.show()In [77]:
from collections import defaultdict
from concurrent.futures import ProcessPoolExecutor, as_completed
import matplotlib.pyplot as plt
from scipy.interpolate import interp1d
from scipy.signal import (butter, coherence, correlate, filtfilt, find_peaks,
resample, sosfiltfilt, welch)
def determine_coherence_per_trial(eeg_data, noise_stim_left, noise_stim_right, channel_labels, channel_selection, lags,
sampling_rate=2048, noise_sample_rate=480, wiggle=0,
upsample_noise=True, eeg_start_time=0.0, eeg_end_time=3.2,
noise_start_time=0.0, noise_end_time=2.0,
bandpass_eeg=None, bandpass_noise=None, freq_band=(50, 80)):
num_samples, num_channels = eeg_data.shape
eeg_segment_start = int(eeg_start_time * sampling_rate)
eeg_segment_end = int(eeg_end_time * sampling_rate)
# Select EEG segment
eeg_data_segment = eeg_data[eeg_segment_start:eeg_segment_end, :].copy()
# Select noise segment
noise_segment_start = int(noise_start_time * noise_sample_rate)
noise_segment_end = int(noise_end_time * noise_sample_rate)
noise_stim_left_segment = noise_stim_left[noise_segment_start:noise_segment_end].copy()
noise_stim_right_segment = noise_stim_right[noise_segment_start:noise_segment_end].copy()
# Optionally bandpass filter the signals
if bandpass_eeg:
lowcut, highcut = bandpass_eeg
for channel in range(num_channels):
eeg_data_segment[:, channel] = bandpass_filter_sos(eeg_data_segment[:, channel], lowcut, highcut, sampling_rate)
if bandpass_noise:
lowcut, highcut = bandpass_noise
noise_stim_left_segment = bandpass_filter_sos(noise_stim_left_segment, lowcut, highcut, noise_sample_rate)
noise_stim_right_segment = bandpass_filter_sos(noise_stim_right_segment, lowcut, highcut, noise_sample_rate)
# Upsample or downsample the noise signals
if upsample_noise:
noise_stim_left_segment = upsample_or_downsample(noise_stim_left_segment, noise_sample_rate, sampling_rate)
noise_stim_right_segment = upsample_or_downsample(noise_stim_right_segment, noise_sample_rate, sampling_rate)
else:
eeg_data_segment = upsample_or_downsample(eeg_data_segment, sampling_rate, noise_sample_rate)
sampling_rate = noise_sample_rate
# Normalise the signals before calculating coherence
noise_stim_left_segment = normalise_signal(noise_stim_left_segment)
noise_stim_right_segment = normalise_signal(noise_stim_right_segment)
eeg_data_segment = normalise_signal(eeg_data_segment)
left_coherence = defaultdict(list)
right_coherence = defaultdict(list)
for channel, channel_label in enumerate(channel_labels):
if channel_label not in channel_selection:
continue
base_lag = lags[channel_label]
best_lag = base_lag
best_coherence_sum = -np.inf
# Check coherence sums within the wiggle room
for delta in range(-wiggle, wiggle + 1):
current_lag = base_lag + delta
eeg_aligned = eeg_data_segment[:, channel][current_lag:current_lag + len(noise_stim_left_segment)]
delay = int(sampling_rate * 1)
f, coh_left = coherence(eeg_aligned[delay:len(eeg_aligned)], noise_stim_left_segment[delay:len(eeg_aligned)], fs=sampling_rate, nperseg=1024)
f, coh_right = coherence(eeg_aligned[delay:len(eeg_aligned)], noise_stim_right_segment[delay:len(eeg_aligned)], fs=sampling_rate, nperseg=1024)
freq_indices = np.where((f >= freq_band[0]) & (f <= freq_band[1]))[0]
coh_left_band = np.mean(coh_left[freq_indices], axis=0)
coh_right_band = np.mean(coh_right[freq_indices], axis=0)
coherence_sum = np.sum(coh_left_band) + np.sum(coh_right_band)
if coherence_sum > best_coherence_sum:
best_coherence_sum = coherence_sum
best_lag = current_lag
# Recompute coherence with the best lag
eeg_aligned_best = eeg_data_segment[:, channel][best_lag:best_lag + len(noise_stim_left_segment)]
delay = int(sampling_rate * 1)
f, coh_left_best = coherence(eeg_aligned_best[delay:len(eeg_aligned_best)], noise_stim_left_segment[delay:len(eeg_aligned_best)], fs=sampling_rate, nperseg=1024)
f, coh_right_best = coherence(eeg_aligned_best[delay:len(eeg_aligned_best)], noise_stim_right_segment[delay:len(eeg_aligned_best)], fs=sampling_rate, nperseg=1024)
left_coherence[channel_label].append(coh_left_best)
right_coherence[channel_label].append(coh_right_best)
return left_coherence, right_coherence, f
def aggregate_and_plot_coherence(experiments, channel_labels, lags, sampling_rate=2048, noise_sample_rate=480, plot_channels=None):
aggregated_left_cued = defaultdict(list)
aggregated_left_uncued = defaultdict(list)
aggregated_right_cued = defaultdict(list)
aggregated_right_uncued = defaultdict(list)
freqs = None
for _, experiment in experiments.iterrows():
eeg_data = experiment['eeg']
noise_stim_left = experiment['noise_stim_left']
noise_stim_right = experiment['noise_stim_right']
attended_side = experiment['attended_side']
left_coherence, right_coherence, freqs = determine_coherence_per_trial(
eeg_data, noise_stim_left, noise_stim_right, channel_labels, plot_channels, lags,
sampling_rate=sampling_rate, noise_sample_rate=noise_sample_rate,
upsample_noise=True, eeg_start_time=0.0, eeg_end_time=3.2,
noise_start_time=0.0, noise_end_time=2.15,
)
if attended_side == 0:
for channel_label in channel_labels:
aggregated_left_cued[channel_label].append(left_coherence[channel_label])
aggregated_right_uncued[channel_label].append(right_coherence[channel_label])
else:
for channel_label in channel_labels:
aggregated_right_cued[channel_label].append(right_coherence[channel_label])
aggregated_left_uncued[channel_label].append(left_coherence[channel_label])
# Plot coherence
if plot_channels is None:
plot_channels = channel_labels
for channel_label in plot_channels:
mean_left_cued = np.squeeze(np.mean(aggregated_left_cued[channel_label], axis=0))
mean_left_uncued = np.squeeze(np.mean(aggregated_left_uncued[channel_label], axis=0))
mean_right_cued = np.squeeze(np.mean(aggregated_right_cued[channel_label], axis=0))
mean_right_uncued = np.squeeze(np.mean(aggregated_right_uncued[channel_label], axis=0))
std_left_cued = np.squeeze(np.std(aggregated_left_cued[channel_label], axis=0))
std_left_uncued = np.squeeze(np.std(aggregated_left_uncued[channel_label], axis=0))
std_right_cued = np.squeeze(np.std(aggregated_right_cued[channel_label], axis=0))
std_right_uncued = np.squeeze(np.std(aggregated_right_uncued[channel_label], axis=0))
fig, ax = plt.subplots(1, 2, figsize=(colwidth, 4))
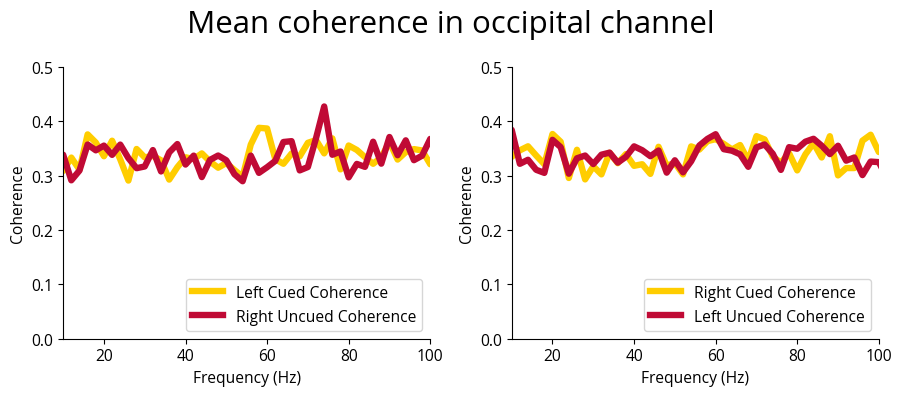
fig.suptitle(f'Mean coherence in occipital channel', fontsize=font_size)
# Left cued and uncued plota
# ax = plt.subplot(1, 2, 1)
ax[0].plot(freqs, mean_left_cued, linewidth=linewidth, color=col_gold, label='Left Cued Coherence')
# ax[0].fill_between(freqs, mean_left_cued - std_left_cued, mean_left_cued + std_left_cued, color=col_gold, alpha=0.2)
ax[0].plot(freqs, mean_right_uncued, linewidth=linewidth, color=col_red, label='Right Uncued Coherence')
# ax[0].fill_between(freqs, mean_right_uncued - std_right_uncued, mean_right_uncued + std_right_uncued, color=col_red, alpha=0.2)
# ax[0].title(f'Coherence for Channel {channel_label} - Left Cued')
# set label
# ax[0].xlabel('Frequency (Hz)')
# ax[0].ylabel('Coherence')
ax[0].spines[['right', 'top']].set_visible(False)
ax[0].legend(loc='lower right', fontsize=font_size/2)
ax[0].set_ylim(0, .5)
ax[0].set_xlim(10, 100)
ax[0].set_xlabel('Frequency (Hz)', fontsize=font_size/2)
ax[0].set_ylabel('Coherence', fontsize=font_size/2)
ax[0].xaxis.set_tick_params(labelsize=font_size/2)
ax[0].yaxis.set_tick_params(labelsize=font_size/2)
# Right cued and uncued plot
plt.plot(freqs, mean_right_cued, linewidth=linewidth, color=col_gold, label='Right Cued Coherence')
# plt.fill_between(freqs, mean_right_cued - std_right_cued, mean_right_cued + std_right_cued, color=col_gold, alpha=0.2)
plt.plot(freqs, mean_left_uncued, linewidth=linewidth, color=col_red, label='Left Uncued Coherence')
# plt.fill_between(freqs, mean_left_uncued - std_left_uncued, mean_left_uncued + std_left_uncued, color=col_red, alpha=0.2)
# plt.title(f'Coherence for Channel {channel_label} - Right Cued')
ax[1].spines[['right', 'top']].set_visible(False)
ax[1].legend(loc='lower right', fontsize=font_size/2)
ax[1].set_ylim(0, .5)
ax[1].set_xlim(10, 100)
ax[1].set_xlabel('Frequency (Hz)', fontsize=font_size/2)
ax[1].set_ylabel('Coherence', fontsize=font_size/2)
ax[1].xaxis.set_tick_params(labelsize=font_size/2)
ax[1].yaxis.set_tick_params(labelsize=font_size/2)
plt.subplots_adjust(left=0, right=1)
plt.tight_layout()
plt.savefig(f'../presentations/figures/avg_coherence.svg', bbox_inches=None, pad_inches=None, transparent=True)
plt.show()
# Example usage
noise_experiments = experiment_data[experiment_data['has_noise']]
lags = {}
for channel_corrs, channel_label in zip(cross_corrs_all_trials, channel_labels):
cumulative_corr = np.sum(channel_corrs, axis=0)
lag = np.argmax(cumulative_corr)
lags[channel_label] = lag
aggregate_and_plot_coherence(noise_experiments, channel_labels, lags, plot_channels=['Iz'])In [25]:
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import cm
import matplotlib.colors as mcolors
import svgwrite
from svgwrite import cm, mm
def create_vector_gabor_patch(size=256, frequency=10, theta=0, sigma=20, phase=0, filename='vector_gabor_patch.svg'):
"""
Generate and save a vector Gabor patch as an SVG file.
Parameters:
size (int): Size of the patch (size x size).
frequency (float): Frequency of the sinusoidal grating.
theta (float): Orientation of the grating in degrees.
sigma (float): Standard deviation of the Gaussian envelope.
phase (float): Phase offset of the sinusoidal grating.
filename (str): Filename to save the SVG file.
"""
# Calculate parameters
theta_rad = np.deg2rad(theta)
half_size = size / 2
# Create SVG drawing
dwg = svgwrite.Drawing(filename, profile='tiny', size=(size * mm, size * mm))
# Define the grid
x = np.linspace(-half_size, half_size, size)
y = np.linspace(-half_size, half_size, size)
X, Y = np.meshgrid(x, y)
# Rotate the grid
X_theta = X * np.cos(theta_rad) + Y * np.sin(theta_rad)
# Create the Gabor function (without the Gaussian envelope for vector representation)
gabor = np.sin(2 * np.pi * frequency * X_theta + phase)
# Normalize to range [0, 1]
gabor_normalized = (gabor - gabor.min()) / (gabor.max() - gabor.min())
# Draw Gabor lines
for i in range(size):
for j in range(size - 1):
# Calculate line intensity and color
intensity = gabor_normalized[i, j]
color = svgwrite.rgb(255 * intensity, 255 * intensity, 255 * intensity, '%')
dwg.add(dwg.line(
start=(j * mm, i * mm),
end=((j + 1) * mm, i * mm),
stroke=color
))
# Save SVG file
dwg.save()
# Generate and save the Gabor patch
create_vector_gabor_patch(size=256, frequency=10, theta=45, sigma=30, phase=0, filename='../presentations/figures/gabor_patch.svg')ModuleNotFoundError: No module named 'svgwrite'